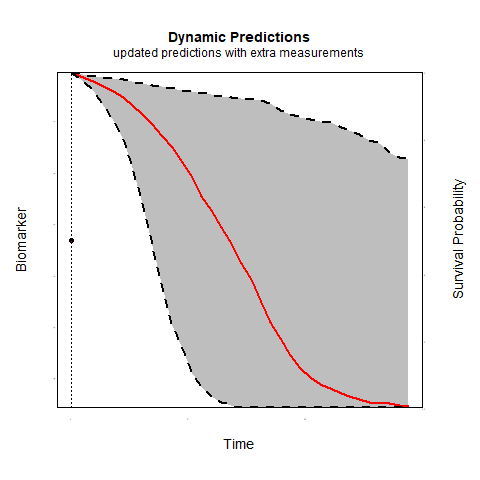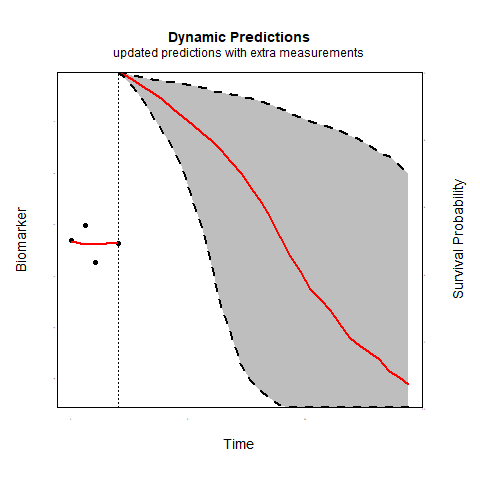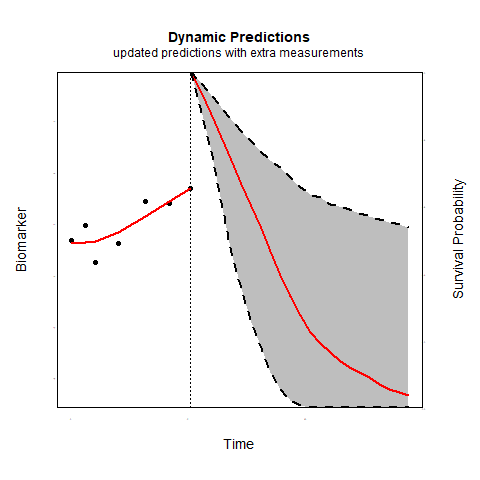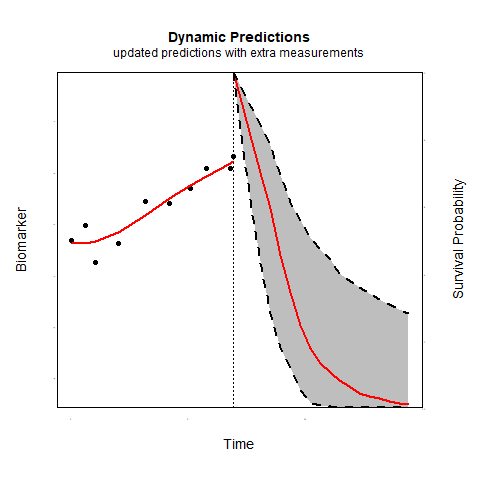
Individualized predictions play a key role in precision medicine and shared decision making. Joint models for longitudinal and survival data have been shown to be a valuable tool in this context.
In this project we study and explore different types of extensions of joint models that can improve the quality of the derived predictions. Some of our proposals are
Time-varying association parameters: Commonly in joint models it is assumed that the coefficients that capture the association between the longitudinal and survival outcomes are constant in time. However, for many biomarkers it is reasonable to assume that their association with the risk of an event may change as patient progress. Using P-splines methodology we investigate whether allowing for time-varying association parameters may improve predictions. The first results indicate that allowing for time-varying effects when in reality the association parameters are constant does not substantially affect predictive performance. However, when the true association structure varies in time, allowing for time-varying parameters considerably outperforms the constant association structure.
Time-varying treatments and invasive procedures: For many conditions in which joint models are used, physicians depending on the progression of patients may choose to change treatments and/or require invasive procedures. In these settings it is highly important to select the optimal timing of such procedures and predict the future outcome of the patient if he would underwent the invasive procedure / change in treatment or not. Motivated by these settings, we develop extensions of joint models that incorporate such decisions during follow-up. The first results indicate that appropriately modeling the patient-specific longitudinal evolutions of the biomarkers before and after the invasive procedure / change in treatment can considerably improve the accuracy of predictions.
Improving predictions by combining biomarkers: Often physicians are interested in combining several longitudinally measured biomarkers for monitoring the progression of patients. From a modeling perspective, different features of these longitudinal profiles of each biomarker may be associated with the risk of an event. For example, the current value, the current slope, the area under profile, the variability of the profile, etc. To improve predictive performance we combine all biomarkers assuming for each one different functional forms, and suitably penalize for model complexity using shrinkage priors.